
Applications of Computational tools in Drug Design & Discovery - Medicinal Chemistry III B. Pharma 6th Semester
Applications of Computational tools in Drug Design & Discovery
Importance of CADD
• Traditional drug discovery includes random screening, serendipitous discovery and process optimization
• Process takes nearly a decade to complete with an average expense of ~300 million dollar
• CADD tends to curtail this expenditure and timeline by providing holistic view
• Evolution of CADD began in 1900s when Emil Fischer (1894) and Paul Ehlrich (1909) propagated the concept of receptors and lock and key mechanisms
• Scientific advancements during the past two decades have changed the way pharmaceutical research generate novel bioactive molecules
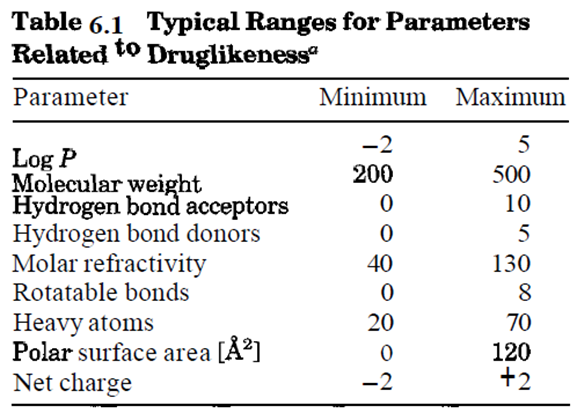
Druglikeness Screening
• Many drug candidates fail in clinical trials because of reasons unrelated to potency against the intended drug target
• Pharmacokinetics and toxicity issues are blamed for more than half of all failures in clinical trials
• First part of virtual screening evaluates the druglikeness of small molecules
• Druglike molecules exhibit favorable absorption, distribution, metabolism, excretion, and toxicological (ADMET) parameters
Discovery of ACE Inhibitor- Captopril
• Angiotensin converting enzyme is a carboxypeptidase with a zinc ion as cofactor
• Plays a key role in the renin-angiotensin cascade involving blood pressure control
• Captopril, a clinically important, potent and reversible inhibitor of ACE
• Design of captopril is one the early endeavours and success of structure based drug design
Information collected for SBDD
• Enzymatic mechanism of ACE was similar to that of carboxypeptidase A
• Exceptional is ACE cleaves off a dipeptide whereas a carboxypeptidase A cleave a single amino acid residue from carboxyl end of a protein
• L-benzylsuccinic acid is a potent inhibitor of carboxypeptidase A
• BPP5a (HOOC-Glu-Lys-Trp-Ala-Pro-NH), a potent pentapeptide inhibitor of ACE that was isolated from the venom of the Brazilian viper
• Captopril is the first ACE inhibitor to enter clinical use following its approval by USFDA in 1981
• Frontline therapeutic agent for the treatment of hypertension and heart failure
Proline
Selectivity or promiscuity? Or both?
• Modern drug development projects should aim to deliver target specific active compounds
• Approach should be- one disease, one target and one drug
• Retrospective analysis proved that approved drugs are promiscuous and bind to several target proteins
• Property of active compound binding to multiple proteins is termed as polypharmacology
• Sorafenib- A Raf inhibitor originally developed against lung or pancreatic cancer
• Proved effective against renal cell cancer by its action on VEGFR2 receptors
• Paxil- A serotonin uptake inhibitor also binds to beta adrenergic receptors offering plausible explanation for increased heart rate
• Prediction of compound polypharmacology has the potential to identify possible adverse effects
• Several methods have been developed for computational prediction of compound polypharmacology
• At present over 5000 drugs, ~10 million virtual library compounds and 1,45,219 biological macromolecule structures are available for exploration which are publicly accessible
• Efficient polyphamacology prediction may be helpful in the future to facilitate drug repurposing and
• To discover more potent drug with less off-target toxicity
Structure of protein
• Have to focus on detailed three dimensional structure of biological molecules
• Like shape or structure of a protein offers clues about the role it plays in the body
• Proteins are shaped to get their job done
• May help in developing new medicines or diagnostic
• Design of lock helps in making key
Proteins are the body’s worker molecules
X-ray Crystallography
Strategies for drug design
• Molecular docking and Dynamics
• Quantitative Structure Activity Relationship
• Pharmacophore modelling
Virtual Screening
• Assessment of overall drug likeness
• Ability to specifically bind to a given drug target
• Goal- reduction of enormous virtual chemical space of small organic molecules to synthesize and/or screen against a specific target to a manageable number of compounds that exhibit the highest chance to lead to drug candidate
• Source of information
• What does a drug look like in general?
• What is known about compounds that interact with the receptor?
• What is known about the structure of target protein and protein-ligand interactions?
• Virtual screening is a category of in silico methods that can be utilized to identify molecules that will (potentially) bind to a target of interest
• These methods are classified as either structure-based or ligand-based:
Before commencing a screen, ask yourself “given the data I have, which virtual screening tool is most appropriate?”
LBDD
• Ligand-Based Drug Design (or indirect drug design):
• Relies on knowledge of other molecules that bind to the biological target of interest
• May be used to derive a pharmacophore model that will define the minimum necessary structural characteristics a molecule must possess in order to bind to the target
SBDD
• Structure-Based Drug Design (or direct drug design):
• Relies on knowledge of the three dimensional structure of the biological target
• Obtained through methods such as X-ray crystallography or NMR spectroscopy
• Using the structure of the biological target, candidate drugs are predicted that will bind with high affinity and selectivity to the target
• Interactive graphics and the intuition of a medicinal chemist are further used in this design process
SP & XP modes of docking
• High Throughput virtual screening (HTVS)
• SP- Standard precision and XP- Extra Precision
• XP scoring function include more stringent terms like hydrophobic effects and charged interactions
• Induced-fit docking (IFD)
• Molecular Dynamics
Chemical compound repositories
| Database | Sample Size |
| PubChem | ~40,000,000 |
| Accerlrys Available Chemicals Directory (ACD) | ~7,000,000 |
| PDBeChem | ~14,572 |
| Zinc | ~21,000,000 |
| LIGAND | ~16,838 |
| DrugBank | ~6711 |
| ChemDB | ~5,000,000 |
| WOMBAT Database | ~331,872 |
| MDDR | ~180,000 |
| 3D MIND | ~100,000 |
Quantitative Structure Activity Relationship
• Most popular approach
• QSAR- computational method to quantify the correlation between chemical structures of series of compounds and a particular chemical or biological process
• Hypothesis behind the concept is similar structural or physicochemical properties have similar activity
Methodology of QSAR
• Identification of ligands with experimentally measured values of desired biological activity.
• Should be of adequate chemically diversity to have large deviation in activity
• Identify and determine molecular descriptors associated with various structural and physico-chemical properties of the molecules under study
Definition of molecular descriptor
• The molecular descriptor is the final result of a logic and mathematical procedure which transforms chemical information encoded within a symbolic representation of a molecule into a useful number, or the result of some standardized experiment
• Roberto Todeschini and Viviana Consonni
What are descriptors?
Includes molecular weight,
Lipophilicity
Hydrogen bonding donors & acceptors
Molecular connectivity
Molecular topology
Molecular geometry
Stereochemistry
Good descriptors should characterize molecular properties important for molecular interactions
Literature suggests that more than 2000 molecular descriptors can be calculated
• Discover correlations between molecular descriptors and the biological activity that can explain the variation in activity in the data set
• Test the statistical stability and predictive power of the QSAR model
• Goal here is to create a molecular “fingerprint” for each molecule that relates to its activity
• Essential part of the drug optimization process
• Success of any QSAR model greatly depends on the
a) choice of molecular descriptors and
b) ability to generate the appropriate mathematical relationship between the descriptors and the biological activity of interest
• Statistical methods applied in QSAR:
a) Multivariable linear regression analysis (MLR)
• Simplest method to quantify the molecular descriptor having good correlation with the variation in activity
• For large numbers of descriptors the MLR method can be time consuming
b) Principle Component Analysis (PCA)
• Efficient method for reduction of the number of independent variables
• Highly useful for systems with a larger number of molecular descriptors than the number of observations
• Results from PCA are often difficult to analyze
c) Partial Least Square analysis (PLS)
• Combination of MLR and PCA techniques
• Gives good correlation
• Advantageous for systems with more than one dependent variable
• Biological systems often display non-linear relationship between the molecular descriptors and the activity
• Once an initial QSAR model has been developed is must be validated
• By internal validation and external validation
Manikanta et al., European Journal of Medicinal Chemistry, 2017, 130, 154-170
Pharmacophore modelling
• Describe 3D features of a molecule
• Molecular descriptors are then combined to create a pharmacophore that can explain the biological activity of the ligands
• A pharmacophore is defined as a spatial arrangement of functional groups and substructures common to active molecules and essential to biological activities
• On the concept of “molecular similarity” of small molecules are derived from a series of active compounds and inactive ones
• Pharmacophore- qualitative aspect
Pharmacophore
• Describe the two or three dimensional arrangement of physicochemical properties of a compound.
• Can be used to screen for molecules with similar arrangement of features.
• In phase , the properties
Ø Include A (acceptor), D (donor), H (hydrophobic), N (Negative), P (positive), and R (aromatic).
Ø Have defined geometry (point, vector or group).
Ø Are defined via SMARTS patterns.
How to create a hypothesis manually
1. Prepare molecule:
· Prepare Ligands (LipPrep)
· Generate Conformers (CobfGen)
2. Create a common hypothesis:
· Align known actives and identify common features
3. Screen a library:
· Identity compounds that match all or most features
· Tweak hypothesis if necessary
Shape based screening
• Hard-sphere overlaps
• Conformers for a screening structure B are aligned to a template structure A.
• Hundreds of trial alignment are considered for each conformer of B.
• The conformer and alignment with the largest A-B overlaps wins.
Conclusion
• CADD- moving drugs from concept to the clinic
• Computational structure-based design supported by medicinal chemistry strategies, can lead to the development of drugs or
• Drug-like molecules with refined pharmacological activity that is better than the parent molecule
• SBDD has been recognized as the tool that facilitated the development of several important drugs in current clinical use or late stage clinical development
• Allowing many drug discovery scientists to carry out more focused, hypothesis-driven discovery initiatives limiting the number of compounds that are synthesized
• Adoption of early stage PK and PD studies has also contributed greatly to the significantly reduced late-stage attrition rate of clinical candidates











0 Comments: