
Transcription
Objectives
At the end of this lecture, students will be able to –
• Discuss the process of transcription in
– Prokaryotes
– Eukaryotes
• Explain post transcriptional modification of mRNA
• Explain the mechanism, functions, and various classes of transcription factors
Content
• Transcription in Prokaryotes
• Transcription in Eukaryotes
Transcription factors
• Sequence specific DNA binding factors
• Protein that binds to specific DNA sequence
• Control the flow of genetic information from DNA to mRNA
• They perform function alone or with other protein in complex
• Act either by promoting or blocking the recruitment of RNA polymerase to specific gene
Protein Synthesis
• Process in which cells build proteins from information in a DNA gene in a two major steps:
I-Transcription and
II-Translation
• Transcription:Synthesis of an RNA (mRNA) that is complementary to one of the strands of DNA
• Translation:Ribosomes read a messenger RNA and make protein according to its instruction
Transcription
• RNA polymerase copies both the exons and the introns.
• Stretch of DNA that is transcribed into an RNA molecule a transcription unit
• A transcription unit contains coding sequence that is translated into protein and sequences that direct and regulate protein synthesis
• Transcription proceeds in the 5' → 3' direction
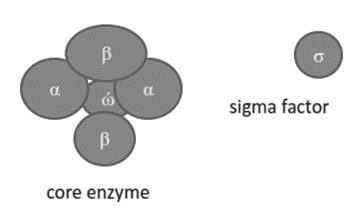
Structure of RNA polymerase
• The holoenzyme is a complete RNA polymerase consisting of an core enzyme and a sigma factor
• The core enzyme consists of 5 polypeptide chains
• Two α subunits, one β and β1 subunit and ώ subunit, σ factor
Functions of RNA polymerase
• Having helicase activity for unwinding.
• Not requires primer
• Lacks proof reading
• σ - Recognition of promoter with the help of transcription factors
• α - activator
• β - phosphodiester bond
• β - DNA template
RNA polymerase
• RNA polymerase;in prokaryotes only the single enzyme
• RNA polymerase governs the synthesis of all cellular RNAs
• In eukaryotic nuclei contain three RNA polymerases
• RNA polymerase I is found in the nucleolus
• The other two polymerases are located in the nucleoplasm
• The three nuclear RNA polymerase have different roles In transcription
• Polymerase I makes a large precursor to the major rRNA (5.8S, 18S and 28S rRNA in vertebrates)
• Polymerase II synthesizes hnRNAs, which are precursors to mRNAs and small nuclear RNAs (snRNAs)
• Polymerase III synthesize the precursor to 5SrRNA, the tRNAs and several other small cellular and viral RNAs
Initiation in prokaryotes
Promoter recognition:
• Sigma factor interacts with core enzyme at β subunit site to check transcription of both the strands by core enzyme
• The holoenzyme transcribes only one of two strands.
• Sigma factor of holoenzyme recognizes the promoter region of the DNA
Promoters in prokaryotes
Centered at -10 to -35 bp from the transcription start point
Promoters in eukaryotes: 3 different promoters
-25 -40 -110
T A T A GC CAAT
• Binding of RNA polymerase: Promoters have binding site for proteins rather than RNA polymerase.
• Most common binding site - a complex of cyclic AMP receptor protein
• Unwinding of DNA double helix: Binding of ώ factor results in unwinding of a double helix
• Open complex allows tight binding of the RNA polymerase with subsequent initiation of RNA synthesis
Synthesis of first base of RNA chain
• The base of RNA synthesized is always in the form of purine i.e. triphosphate guanine (ppp G) or adenine (ppp A)
• Initiation of mRNA synthesis does not require primer Initiation ends after the formation of first inter nucleotide bond.
Elongation
• Core enzyme moves along the template from 3ꞌ-5ꞌ end untwisting the helix bit by bit and adding one complementary nucleotide
• After 8-9 bp of RNA synthesis occurs, sigma factor is released and recycled for other reaction
• RNA polymerase completes the transcription at 30-50 bp/sec
• Unwinding and rewinding of DNA occurs simultaneously
Termination
Two types of terminator sequence occur in prokaryotes
1. Type 1(ρ- independent):
• RNA molecule terminated without the aid of the rho factor contain GC rich sequence followed by U residues
• GC region makes RNA to spontaneously fold into hairpin loop that tends to pull the RNA away from DNA
• The weaker bonds between the sequence of U residues and DNA template broken releasing the newly formed RNA molecule
2. Type 2 (rho- dependent):
• RNA molecule that do not form GC rich hairpin loop requires rho factor for Termination
• It is a hexameric factor which binds to specific termination sequence 50-90 bases located near 3ꞌend of newly forming RNA molecule
• It acts as an ATP – dependent unwinding enzyme, unwinds RNA from DNA template as it proceeds
Transcription in eukaryotes Initiation
• RNA polymerase cannot recognize the promoters, requires general transcription factor (GTFs)
• Promoters forms pre intiation complex with GTFs
• Assembly of the proteins to TATA box forms TATA binding protein (TBP)
• TBP is present as a subunit of much larger protein complex called TFІІD which specially binds to TATA box
Initiation
• TFІІB provide a binding site for RNA polymerase
• TFІІF contains subunit homologous to the bacterialσ factor, bounds to the entering polymerase
• TFІІH contains 10 subunits, 3 possess enzymatic activity helps in unwinding the DNA (helicase activity)
Elongation
• Same as prokaryotes
• Involves sequential addition of nucleotide units
Termination:
• Transcription by RNA polymerase І is terminated by a protein factor that recognizes an 18-nucleotide termination signal
• Termination signals for RNA polymerase III include short run of Us (as in prokaryotic signal). No proteins factors are needed for their recognition
Post transcription processing of mRNA
• Post-transcriptional modification is a process by which, in eukaryotic cells, primary transcript RNA is converted into mature RNA
• Conversion of precursor messenger RNA into mature messenger RNA (mRNA), which includes splicing and occurs prior to protein synthesis
• The pre-mRNA molecule undergoes three main modifications
• 5' capping
• 3' polyadenylation
• RNA splicing - occur in the cell nucleus before the RNA is translated
• The 5' capping:5ꞌ end chemically modified by the addition of 7 methylguanosine
• Replacement of triphosphate group at the 5' end of the RNA chain with a special nucleotide GMP nucleotide
3’ adenylation
• Addition of poly A tail to 3’ end
• Added before it leaves the nucleus
• AAUAA sequence recognized by a specific endonuclease that cleaves the RNA around 20 nucleotide down stream
• Poly A tail associated with protein, retard action of 3’- exonucleases
Splicing
• RNA splicing - introns are removed from the pre-mRNA
• Remaining exons connected to re-form a single continuous molecule
• Catalyzed by a large protein complex called the spliceosome
• Allows production of a large variety of proteins from a limited amount of DNA
Mechanism of transcription factors
• Stabilize or block the binding of RNA polymerase to DNA
• Catalyse acetylation or deacetylation of histone
• Histone acetyl transferase activity – acetylates histone proteins
• Histone deacetylases activity – decetylates histone proteins
• Recruit coactivators or corepressor proteins to the transcription factor-DNA complex
Functions of transcription factors
• Reads and interprets the genetic “blue print” in the DNA
• Bind to DNA and initiate program of increase or decrease gene transcription
• Basal transcription regulation
– General transcription factors necessary for transcription to occur
– They interact with polymerase directly
Functions of transcription factors
Differential enhancement of transcription
• Regulate the expression of various gene by binding to enhancer region of DNA adjacent to regulated gene
• Ensure that genes are expressed in right cell at the right time
Response to intracellular cells
• Transcription factors are involved in the downstream of signalling cascade
Response to environment
• Involved in downstream of signaling cascade in environmental stimuli
• Heat shock factors, hypoxia inducible factors
Cell cycle control
• Proto-oncogenes or tumor suppressor gene regulate cell cycle
• Myc oncogene – in cell growth & apoptosis
Regulation of transcription factors
Synthesis
• Transcription factors are transcribed from gene on a chromosome into RNA, then RNA to proteins
• Any of these steps can be regulated to affect the production of transcription factors
Nuclear localization
• Transcribe in nucleus but not translated in cytoplasm
• They have nuclear localization signals that direct them to nucleus
Activation
• Transcription factors can be activated or inactivated by through signal sensing domain
• Ligand binding -influence factors present in cell; decided if factors are in active state or capable to bind DNA
• Phosphorylation -STAT protein must be phosphorylated before they can bind to DNA
• Interaction with other transcription factors
Accessibility of DNA binding site
• DNA in nucleosome is inaccessible to many transcription factors
• Nucleosome should be actively removed by molecular motors
Availability of other cofactors/ transcription factors
• Most transcription factors do not work alone
• For transcription many factors must bind to DNA regulatory sequence
• Recruitment of intermediary proteins
Classes of transcription factors
Classified based on –
• Mechanism of action
• Regulatory function
• Sequence homology in their DNA binding domain
Mechanistic class of transcription factor
• General transcription factors
– Involved in the formation of pre initiation complex
– TFIIA, TFIIB, TFIID, TFIIE, TFIIF
– Ubiquitous, interact with core promoter region
• Upstream transcription factors
– Binds upstream to initiation site
– Stimulate or repress transcription
Functional class of transcription factors
Constitutively active
• Present in all cells at all time – general transcription factors Sp1, NF1, and CCAAT
Conditionally active – requires activation
• Developmental – cell specific
• Signal – dependent - requires external signal for activation
Structural class of transcription factor
• Based on sequence similarity and tertiary structure of DNA- binding domain
• 1- superclass – Basic domains
• 2 – Superclass – Zinc co-ordinating DNA binding domain
• 3 Superclass – Helix-turn-helix
• 4 superclass- beta-scaffold factors with minor groove contacts
• 5 superclass – other transcription factors
Summary
• Protein synthesis in prokaryotes and eukaryotes occurs through two steps – Transcription and Translation
• Process of transcription involves 3 steps – initiation, elongation and termination
• Process of transciption in similar in prokaryotes and eukaryotes but differs in post transcriptional modification
• Post transcriptional modifications include addition of 5’ cap, 3’ adenylation and RNA splicing
• Transcription factors are sequence specific DNA binding factors and protein that binds to specific DNA sequence
• Stabilize or block the binding of RNA polymerase to DNA
• They reads and interprets the genetic “blue print” in the DNA
• Also bind to DNA and initiate program of increase or decrease gene transcription
• Transcription factor is classified based on mechanism of action, regulatory function and sequence homology











0 Comments: